We take it for granted that in the age of antibiotics there will always be a drug available to treat an infection. But reports of outbreaks of a pan-resistant fungal disease known as Candida auris– that is resistant to every antifungal medication physicians have to treat these pathogens–have brought into focus just how vulnerable we are to the microbes around us (as if the COVID-19 pandemic hadn’t taught us that already).1 Three deaths had been associated with pan-resistant Candida auris by the time the pathogen had received any media coverage, and the number of cases of C. auris has risen steadily ever since.2 Pan resistance signifies the non-susceptibility of a pathogen to all antimicrobial agents commercially available to eliminate the infection. Among hospitalized patients in the United States (US), antifungal-resistant C. auris causes approximately 400 infections, compared with approximately 35,000 infections caused by other antifungal-resistant Candida species.3
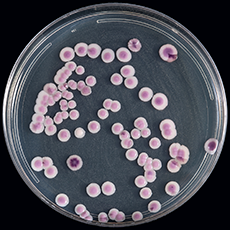
Candida auris on CHROMagar Candida, displaying multiple color morphologies
Source: Identification of Candida auris | Candida auris | Fungal Diseases | CDC
Candida auris first came on the scene in 2009 when it was identified in Asia and has been found to cause infections of the bloodstream, wounds, and ear. Bloodstream infections caused by this organism can have a 30 to 60% mortality rate, even without pan-resistance. This high mortality rate places C. auris near the top to the Center for Disease Control and Prevention (CDC) list of Urgent Threats.4 People in nursing homes and other long-term care facilities, as well as those patients with breathing tubes, feeding tubes, or catheters, seem to be most at risk for C. auris infections. This organism spreads readily through person-to-person contact and by touching contaminated surfaces; it is also difficult to eradicate through normal antimicrobe cleaning protocols. As it easily can be confused with other types of yeasts, specialized laboratory tests are required for proper identification of C. auris. Since its isolation in Japan in 2009, it has spread to 40 countries and was first isolated in the US in 2015. Interestingly, retrospective analysis of yeast collections identified the earliest known C. auris isolate in South Korea in 1996.
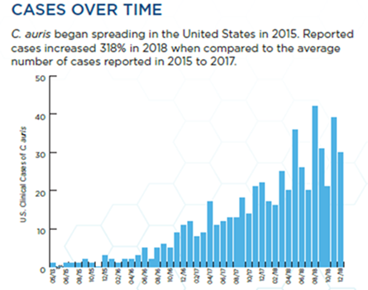
Source: Drug-Resistant Canida Auris (cdc.gov)
Three main classes of antifungal medications are available for treating invasive yeast infections: azoles (such as fluconazole), polyenes (namely amphotericin B) and echinocandins (i.e., caspofungin). In the United States, approximately 85% of C. auris isolates are resistant to azoles, 33% are resistant to amphotericin B, and 1% to echinocandins; pan-resistant isolates are resistant to all three classes of drugs.5
As mentioned above, the alarm was raised in the summer of 2021 with reports that outbreaks of C. auris infections had occurred between January and April in Washington D.C. and Texas. Out of the 101 cases of this infection in Washington D.C., three were caused by pan-resistant isolates. In Texas there were 22 cases of C. auris identified, and two isolates were found to be pan-resistant; another five of the Texas isolates were resistant to two of the three classes of antifungal medications.6 Typically, an isolate from a patient can become resistant to an antimicrobial following treatment with that antimicrobial, but it is notable that in these cases these patients had not been treated with antifungal agents before they became infected, which indicates that the C. auris strain they acquired was already pan-resistant. The low incidence of echinocandin resistance points to the seriousness of the threat of pan-resistance in this organism.
A recent study analyzed the development of resistance to fluconazole in clinical isolates of C. auris to determine how quickly mutations associated with antibiotic resistance arise, how stable those mutations are, and whether there was a fitness cost to the organism.7 Typically, it can take several rounds of organism replication to see resistance to an antibiotic but in this study, resistance at high levels was rapidly observed. Importantly, the investigators saw no cost for the resistant organisms to maintain these mutations, even in the absence of fluconazole—again, an unusual observation as the genes involved in antibiotic resistance are generally involved in a cellular function that is compromised when mutated. Not only did fluconazole resistance arise rapidly, but in some cases the isolates were resistant to very high concentrations of the drug. Perhaps the most striking finding in this study was the identification of a so-called mutator gene (a homolog of the DNA mismatch repair gene) in a clinical isolate of C. auris, which, when altered, could lead to higher mutation rates and could be a factor in the acquisition of antifungal resistance in C. auris.
Since C. auris can often be misidentified as other yeasts (most often as Candida haemulonii, another rare yeast), the proper laboratory methods for identification can be important for patient treatment and infection control measures.8 Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is the most reliable method for identification of C. auris since there are no phenotypic characteristics that distinguish this organism from other Candida species. Other acceptable methods of identification rely on DNA sequencing of the D1 – D2 region of the 28S ribosomal DNA (rDNA), or the internal transcribed spacer (ITS) region of rDNA.
Besides its resistant to antifungal drugs, C. auris seems to possess features out of sync with other Candida species – one of which is its ability to form what are called “dry biofilms.”9 Unlike most biofilms formed by microorganisms, which occur under moist or wet conditions, these can remain on dry surfaces, like hospital bed railings, and spread easily from patient to patient or from patient to caregiver.9 These dry biofilms are very difficult to eliminate. This organism has also adapted well to warm environments, so it is able to cause invasive infections, unlike many yeast species, which are not as thermotolerant. In addition, C. auris has not been detected in nature (plants, insects, or aquatic environments), unlike related species.10
Strikingly, four different strains of C. auris emerged around the same time worldwide during 2012 – 2015, and all four have been found in the US. Isolates were collected from 54 patients with C. auris infections from Pakistan, India, South Africa, and Venezuela during that time, and susceptibility testing and whole genome sequencing (WGS) was conducted.11 Four percent of the isolates were resistant to all three antifungal classes, 93% were resistant to fluconazole, 35% to amphotericin B, and 7% to echinocandins; 41% were resistant to two antifungal classes. Following WGS the isolates could be grouped into unique clades based on geographic region; the clades could be separated by thousands of single-nucleotide changes but within each clade the isolates were found to be clonal. This analysis indicates that these clones of C. auris emerged nearly simultaneously and independently on three continents – South Africa, South Asia, and South America.10
The success of C. auris as a human pathogen in such a broad and rapid fashion underscores the urgent menace that microbes can pose for us. This organism also presents community and healthcare infection control threats that must be considered, and so will likely keep us on our toes for the foreseeable future.
Sources
1. Jacobs, A. “C.D.C. Warns of Superbug Fungus Outbreaks in 2 Cities.” The New York Times. https://www.nytimes.com/2021/07/23/health/superbug-fungus-cdc.html.
2. “Weekly cases of notifiable diseases, United States, U.S. territories, and Non-U.S. Residents week ending August 20, 2022 (Week 33) Table1f Brucellosis; Campylobacteriosis; Candida auris, clinical.” CDC. https://wonder.cdc.gov/nndss/static/2022/33/2022-33-table1f.html.
3. “CDC Actions to Prevent the Spread of Antifungal Resistance.” CDC. https://www.cdc.gov/fungal/pdf/cdc-antifungal-resistance-508.pdf.
4. “Antibiotic Resistance Threats in the United States, 2019.” CDC. https://www.cdc.gov/drugresistance/biggest-threats.html.
5. “Notes from the Field: Transmission of Pan-Resistant and Echinocandin-Resistant Candida auris in Health Care Facilities ― Texas and the District of Columbia, January–April 2021.” CDC. https://www.cdc.gov/mmwr/volumes/70/wr/mm7029a2.htm.
6. Gamillo, E. “CDC Reports Several Cases of Drug-Resistant Fungal Infection in Two U.S. Cities.” Smithsonian Magazine. https://www.smithsonianmag.com/smart-news/cdc-reports-outbreaks-drug-resistant-fungal-infection-two-us-cities-180978317/.
7. Burrack, L. S., R. T. Todd, N. Soisangwan, N. P. Wiederhold, A. Selmecki. “Genomic Diversity across Candida auris Clinical Isolates Shapes Rapid Development of Antifungal Resistance In Vitro and In Vivo.” PubMed. https://pubmed.ncbi.nlm.nih.gov/35862787/.
8. “Candia auris.” CDC. https://www.cdc.gov/fungal/candida-auris/fact-sheets/fact-sheet-lab-staff.html.
9. Ahmad, S and Alfouzan, W. 2021. Candida auris: Epidemiology, diagnosis, pathogenesis, antifungal susceptibility, and infection control measures to combat the spread of infections in healthcare facilities. Microorganisms. 9:807. https://doi.org/10.3390/microorganisms9040807
10. Casadevall A., D. P. Kontoyiannis, V. Robert. “On the Emergence of Candida auris: Climate Change, Azoles, Swamps, and Birds.” ASM. https://journals.asm.org/doi/10.1128/mBio.01397-19.
11. Lockhart, S. R., et al. “Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses.” NLM. https://pubmed.ncbi.nlm.nih.gov/27988485/.





0 Comments